Publication and release
Overview
Teaching: 20 min
Exercises: 10 minQuestions
Why should I make my research objects available?
What open source tools to use for applying data science practices in bioscience?
How to get your research work cited and invite more contributions to your project?
Objectives
First learning objective. (FIXME)
Publications

While the output of research projects is usually centred around publishing a journal article, this format of science communication and knowledge sharing is increasingly restrictive with the new ways scientific research is conducted. The requirements from journals themselves is also expanding, you are now often asked to upload data sets and code as part of your publication. Releasing data is increasingly a requirement from funding bodies, and outputs from research groups can go beyond a single paper, releasing tools and methods that can be used worldwide.
In general there are different degrees of openness.
- Fully private data and code, unavailable
- Pseudo-open – “available on request”
- Released static code parallel to a research paper, see Zenodo or Figshare
- Open online repository – CRAN, GitHub
- Collaborative, open science tool with ongoing development
What can be released:
- Open Data: Documenting and sharing research data openly for re-use.
- Open Source Software: Documenting research code and routines, and making them freely accessible and available.
- Open Hardware: Documenting designs, materials, and other relevant information related to hardware, and making them freely accessible and available.
- Open Access: Making all published outputs freely accessible for maximum use and impact.
- Open Notebooks: An emerging practice, documenting and sharing the experimental process of trial and error.
https://the-turing-way.netlify.app/reproducible-research/open.html
Open or Private?
Researchers often worry that they need to hide their code to prevent others stealing it.
“After giving talks about open science I’ve sometimes been approached by skeptics who say, ‘Why would I help out my competitors by sharing ideas and data on these new websites? Isn’t that just inviting other people to steal my data, or to scoop me? Only someone naive could think this will ever be widespread.’ As things currently stand, there’s a lot of truth to this point of view. But it’s also important to understand its limits. What these skeptics forget is that they already freely share their ideas and discoveries, whenever they publish papers describing their own scientific work. They’re so stuck inside the citation-measurement-reward system for papers that they view it as a natural law, and forget that it’s socially constructed. It’s an agreement. And because it’s a social agreement, that agreement can be changed. All that’s needed for open science to succeed is for the sharing of scientific knowledge in new media to carry the same kind of cachet that papers do today”
Nielsen, M. Reinventing Discovery: The New Era of Networked Science. Princeton University Press, 2011.
https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000246
Code release
For computational projects, releasing your work in an open repository has parallels with publications.

There can be specific requirements to keep code bases and/or data private. See the section below for good and not so good reasons for keeping work private.
You can release code and data associated with a research article as a series of files/folders. If your project follows the folder template introduced in a previous episode, for example:

Examples of a template folder tree for a computational project. https://github.com/tonic-team/Tonic-Research-Project-Template
You could bundle folders into a .zip file and upload it to Zenodo.
Zenodo

Zenodo is a general-purpose open-access repository developed under the European OpenAIRE program and operated by CERN. It allows researchers to deposit research papers, data sets, research software, reports, and any other research related digital artefacts.
Uploads to Zenodo are:
- Safe — your research is stored safely for the future in CERN’s Data Centre for as long as CERN exists.
- Trusted — built and operated by CERN and OpenAIRE to ensure that everyone can join in Open Science.
- Citeable — every upload is assigned a Digital Object Identifier (DOI), to make them citable and trackable. No waiting time — Uploads are made available online as soon as you hit publish, and your DOI is registered within seconds.
- Open or closed — Share e.g. anonymized clinical trial data with only medical professionals via our restricted access mode.
- Versioning — Easily update your dataset with our versioning feature.
- GitHub integration — Easily preserve your GitHub repository in Zenodo.
- Usage statistics — All uploads display standards compliant usage statistics
Citable Code
The Citation File Format provides citation metadata, for software or datasets, in plaintext files that are easy to read by both humans and machines.
Adding a CITATION.cff file to your folder means it can be cited when others use it, increasing recognition for your work and your research project’s impact.
See more at The Turing Way: CITATION.cff
https://the-turing-way.netlify.app/_images/software-credit.jpg
Collaborative Open Code

Downloading code and data files from Zenodo or other open access repositories can be useful when someone wants to review your the final outcome of your computational work. However, with an open GitHub repository, sharing code becomes much more collaborative and in real-time.

Uploading code in progress to an open GitHub Repo is the best and most well-used method for programming collaboration.
As you develop a tool or methodology, users have the ability to use your code while it is a work in progress and others can contribute or add features.

When using specifically R, you could release R packages on CRAN where anyone can then download and use you code.
Open Science Tools – Research Software with Impact
Many research groups produce widely used tools and software that are used across biomedical and life sciences. Examples of an open science tool in ongoing development and collaboration:
DeepLabCut
https://github.com/DeepLabCut/DeepLabCut
A toolbox for markerless pose estimation of animals performing various tasks.

Cellpose
https://github.com/MouseLand/cellpose
https://cellpose.readthedocs.io/en/latest/
A generalist algorithm for cell and nucleus segmentation.

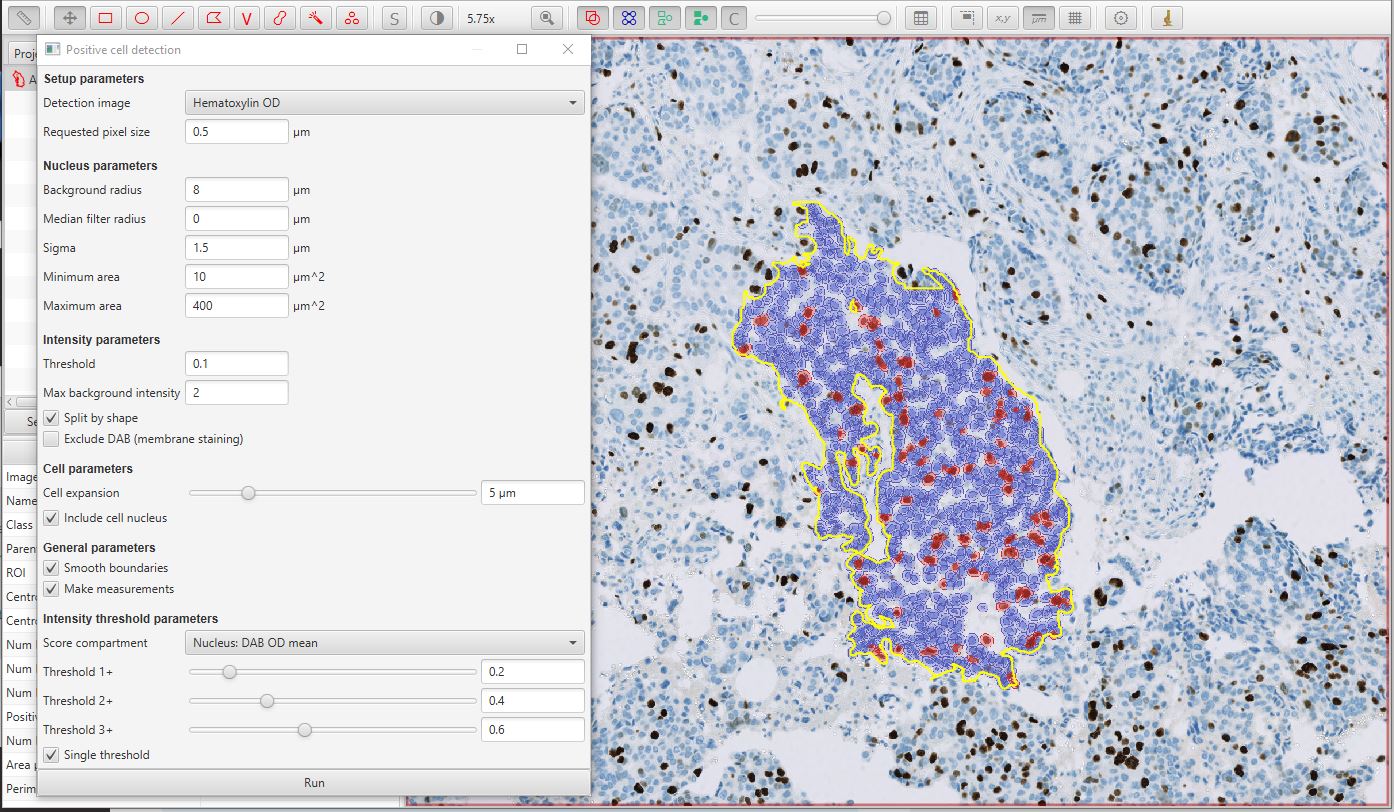
Qupath
https://github.com/qupath/qupath
Extensive tools to annotate and view images, including whole slide & microscopy images. Interactive machine learning for both object & pixel classification.
Research Objects can be Released with Digital Object Identifiers (DOI)
DOIs are alphanumerical unique and persistent identifiers with a permanent web address for different research objects that can be cited by you and other researchers. Each pre-print and publication is published with a DOI, but independent of the paper, different research objects can be published online on servers that offer DOIs at any stage of your research. Some of these servers are Zenodo, FigShare, Data Dryad (for data), Open Grants (for grant proposals) and Open Science Framework (OSF) (for different components of an open research project). It allows you to show connections between different parts of research as well as cite different objects from your work independently.
When working on GitHub for instance, you can connect the project repository with Zenodo to get a DOI for your repository. The Citation File Format, then lets you provide citation metadata, for software or datasets, in plaintext files that are easy to read by both humans and machines. Read the Making Research Objects Citable chapter in The Turing Way Guide to Communication.
Key Points
First key point. Brief Answer to questions. (FIXME)